1. StereoHub
1.1 StereoHub Architecture:
StereoHub: a comprehensive cloud platform for Stereo-Seq analysis, Single-Cell visualization, and CT segments annotation.
1. Github Team: https://github.com/StereoHub/
2. Source Repository: https://github.com/StereoHub/StereoHub
3. Documents: https://stereohub.github.io
4. Cloud Platform: http://43.242.96.52:5002
1.2 StereoHub Features:
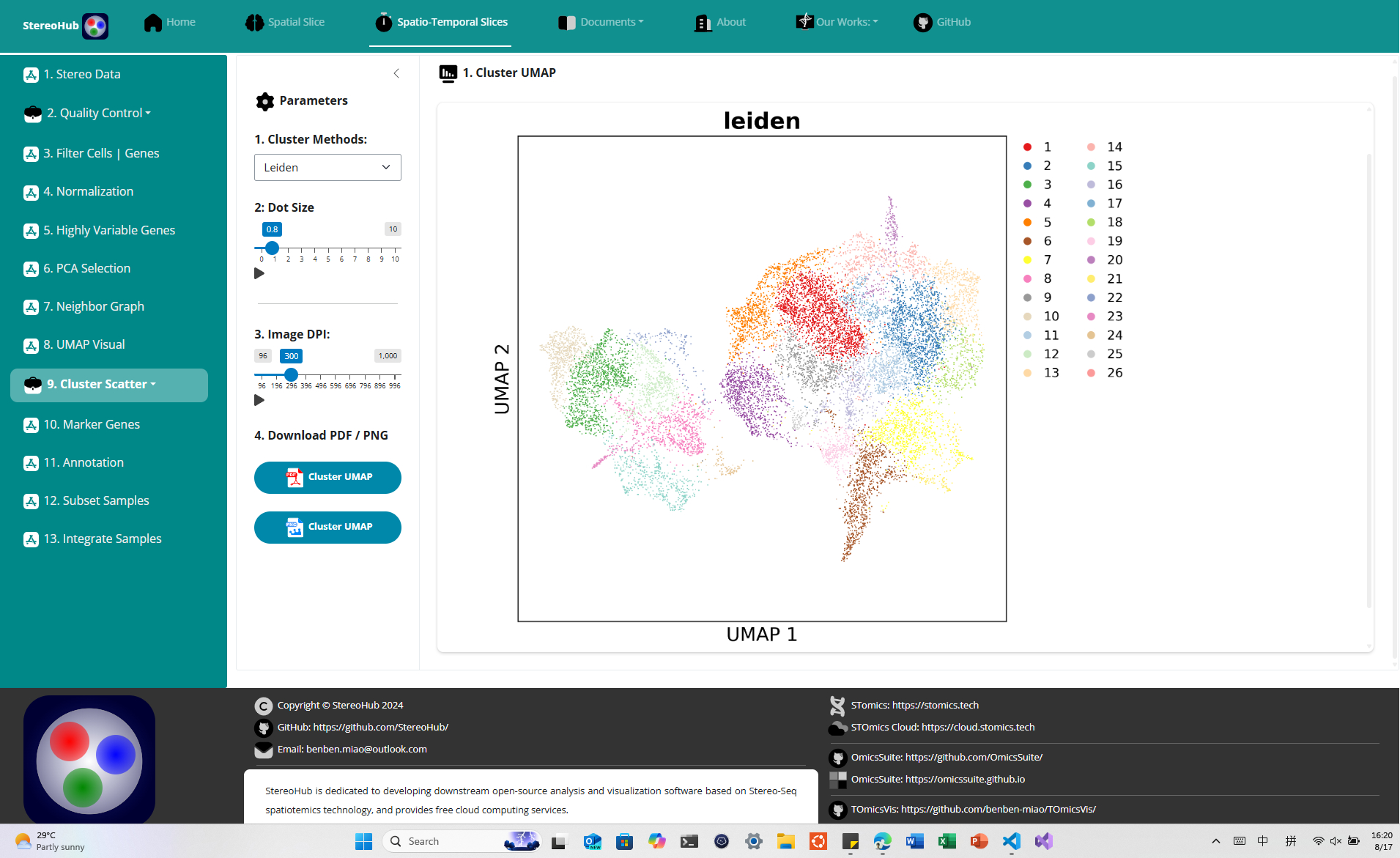
1.2.1 Stereo-Seq Analysis
Pipelined single-slice analysis and time-based multi-slice analysis.
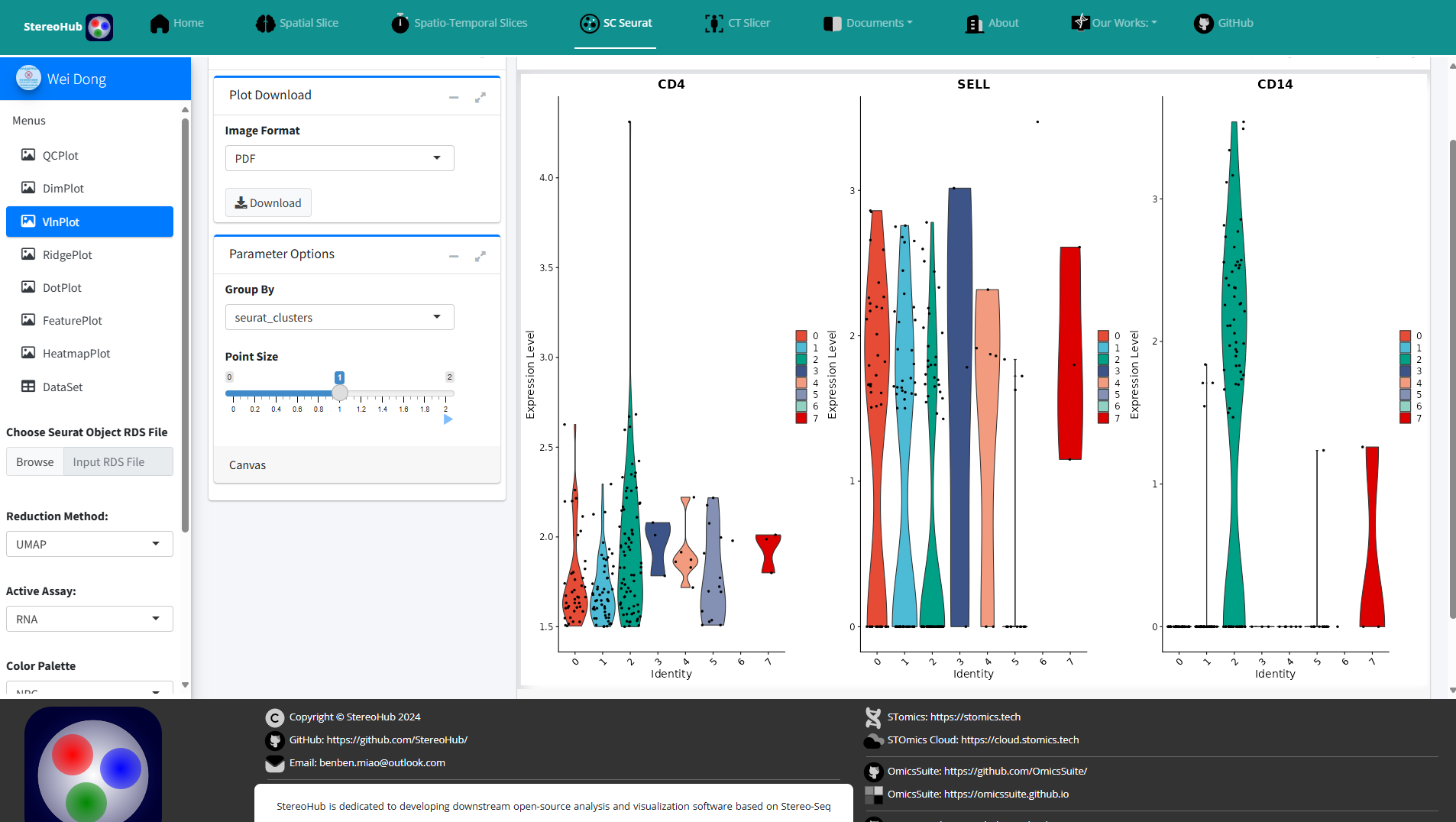
1.2.2 Single-Cell Seurat
Pipelined analysis and visualization of Single-Cell expression data based on Seurat.
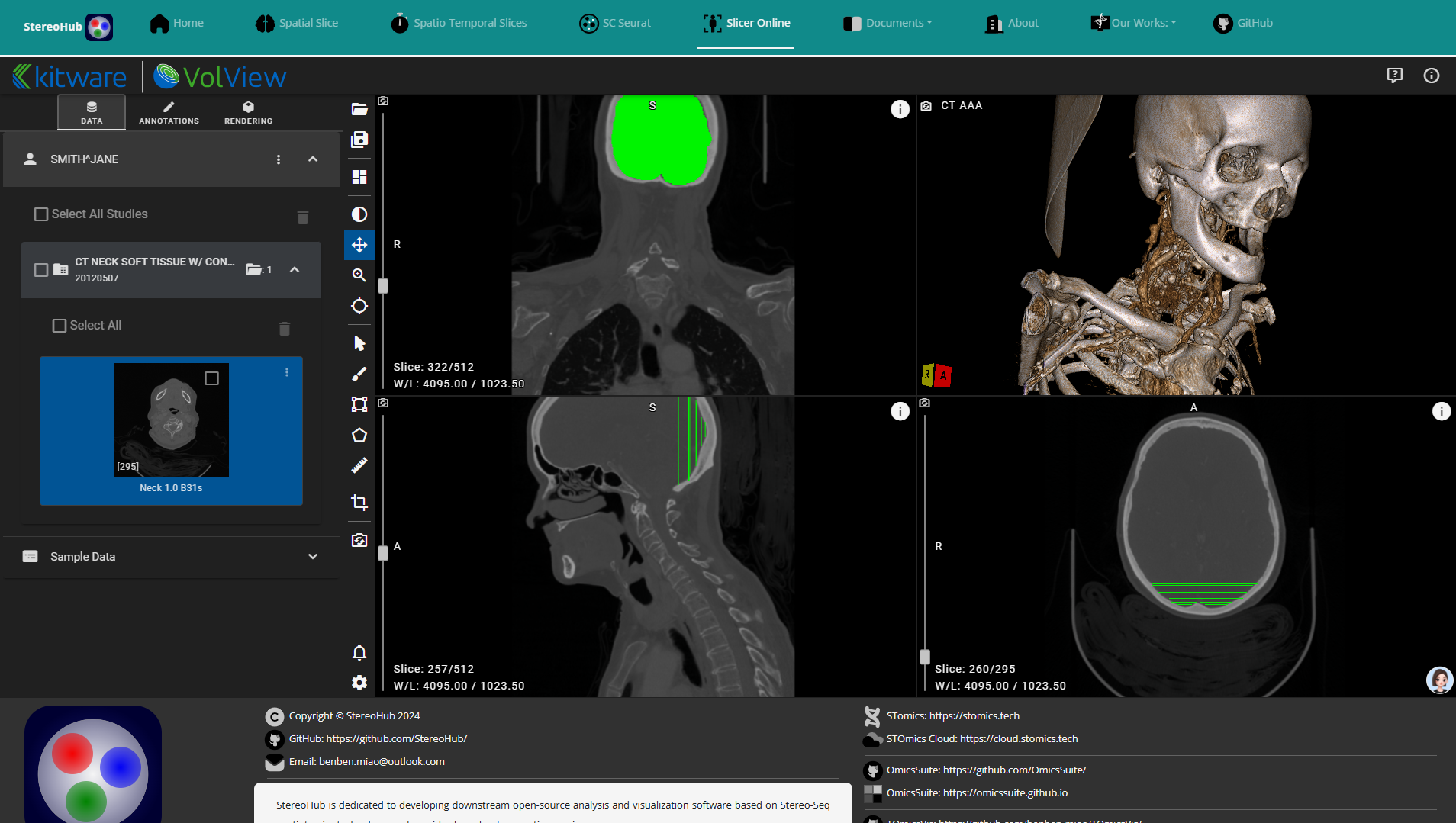
1.2.3 Computed Tomography Slicer
Supports online labeling and feature extraction of DICOM data from CT and NMR.
1.3 StereoHub Develop or Deploy
1.3.1 Installing
# 1. git clone repository
git clone https://github.com/StereoHub/StereoHub.git
cd StereoHub
# 2. Install Environment
# 2.1 For Windows (Recommended)
.\env-win.bat
# OR
# 2.2 For Linux (Recommended)
bash env-linux.sh
# OR
# 2.3 Install Steps (Recommended)
# 2.3.1 Mamba Env Create and Activate
mamba create -n stereohub python=3.8
mamba activate stereohub
# 2.3.2 Shiny
mamba install -c conda-forge shiny=1.0.0 shinywidgets=0.3.2 IPython=8.12.2 ipywidgets=8.1.3
# 2.3.3 Utils
mamba install -c conda-forge numpy=1.23.5 pandas=1.5.3 matplotlib=3.7.1 faicons=0.2.2
# 2.3.4 Stereopy
mamba install stereopy=1.3.1 -c stereopy -c grst -c numba -c conda-forge -c bioconda -c fastai -c defaults
# OR
# 2.4 Conda Env Export and Create (Not Recommended)
conda env create -f stereohub.yml
conda activate stereohub
1.3.2 Run, develop and debug
# 1. For Windows
.\start-win.bat
# 2. For Linux
bash start-linux.sh
# 3. All Terminals
python -m shiny run \
--host 127.0.0.1 \
--port 5000 \
--reload \
--reload-includes "*.py,*.css,*.js,*.html,*.md" \
--reload-excludes "*.png,*.pdf" \
--log-level info \
--app-dir "." \
--launch-browser \
--dev-mode \
app.py
1.3.3 Deploy: Shinylive (WebAssembly + Pyodide) (Not Recommended)
Shinylive: Shiny + WebAssembly: https://shiny.posit.co/py/docs/shinylive.html
shiny create myapp
pip install shinylive
shinylive export myapp site
python3 -m http.server --directory site 5000
1.3.4 Deploy: Shiny-Server (Recommended)
Self-hosted deployments: https://shiny.posit.co/py/docs/deploy-on-prem.html
# 1. shiny-server
wget https://download3.rstudio.org/centos7/x86_64/shiny-server-1.5.22.1017-x86_64.rpm
sudo rpm -ivh shiny-server-1.5.22.1017-x86_64.rpm
# 2. stereohub environment
bash env-linux.sh
# 3. shiny-server home
cd /srv/shiny-server/
git clone https://github.com/StereoHub/StereoHub.git
# 4. shiny-server config
sudo vim /etc/shiny-server/shiny-server.conf
python /cluster/envs/miniconda3/envs/stereohub/bin/python;
run_as shiny;
server {
listen 3838;
# Define a location at the base URL
location / {
# Host the directory of Shiny Apps stored in this directory
site_dir /srv/shiny-server;
# Log all Shiny output to files in this directory
log_dir /var/log/shiny-server;
# An index of the applications available in this directory will be shown.
directory_index on;
}
}
# 5. shiny-server manage
sudo systemctl start shiny-server
sudo systemctl status shiny-server
sudo systemctl restart shiny-server
1.3.5 Deploy: Docker (Recommended)
# 1. Build, Run, Push
# 1.1 Setting services name, build, tag, port, volume, etc.
vim docker-compose.yml
# 1.2 Build stereohub and run container
docker compose up stereohub -d --build
# 1.3 Push stereohub
docker push omicsdocker/stereohub:1.1.0
# 2. Pull from DockerHub
# 2.1 Pull image with tag
docker pull omicsdocker/stereohub:1.1.0
# 2.2 Run service with port and name
docker run -d -p 3838:3838 --name stereohub omicsdocker/stereohub:1.1.0
1.3.6 Deploy: Singularity (Recommended)
# 1. Build Singularity Image File
# 1.1 Build based Docker local image
singularity build stereohub.sif docker-daemon://omicsdocker/stereohub:1.1.0
# 1.2 Build based Docker remote image
singularity build stereohub.sif docker://omicsdocker/stereohub:1.0.0
# 1.3 Build based singularity.def
singularity build stereohub.sif singularity.def
# 1.4 Run SIF
singularity run --bind /var/log/shiny-server:/var/log/shiny-server stereohub.sif
sudo singularity run --writable-tmpfs --bind /var/log/shiny-server:/var/log/shiny-server stereohub.sif
2. Stereo-Seq Technology:
a. STomics Stereo-Seq: https://stomics.tech
b. STomics Cloud: https://cloud.stomics.tech
c. STOmics Database: https://db.cngb.org/stomics/
d. ImageStudio, StereoMap: https://stomics.tech/products/BioinfoTools/OfflineSoftware
e. STomics Github: https://github.com/STOmics/
f. Stereopy: https://github.com/STOmics/Stereopy/
2.1 References
Ao Chen, Sha Liao, Mengnan Cheng, Longqi Liu, Xun Xu, Jian Wang. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell, 2022, doi: https://doi.org/10.1016/j.cell.2022.04.003